-Search query
-Search result
Showing 1 - 50 of 805 items for (author: bak & j)

EMDB-42542: 
CryoEM Structure of Allosterically Switchable De Novo Protein sr322, In Closed State without Effector Peptide, off Target Multimeric State
Method: single particle / : Weidle C, Borst A

EMDB-42442: 
CryoEM Structure of Allosterically Switchable De Novo Protein sr322, In Closed State without Effector Peptide
Method: single particle / : Weidle C, Borst A

EMDB-42491: 
CryoEM Structure of Allosterically Switchable De Novo Protein sr312, in Open State with Effector Peptide
Method: single particle / : Weidle C, Skotheim R
EMDB-43516: 
Cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

EMDB-43517: 
Cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

PDB-8vt2: 
cryo-EM structure of HMPV (MPV-2c)
Method: single particle / : Yu X, Langedijk JPM

PDB-8vt3: 
cryo-EM structure of HMPV (MPV-2cREKR)
Method: single particle / : Yu X, Langedijk JPM

EMDB-43551: 
CCHFV GP38 bound with ADI-46143 and ADI-46158 Fabs
Method: single particle / : Hjorth CK, McLellan JS

EMDB-43552: 
CCHFV GP38 bound with ADI-58062 and ADI-63530 Fabs
Method: single particle / : Hjorth CK, McLellan JS

EMDB-43553: 
CCHFV GP38 bound with ADI-58026 and ADI-63547 Fabs
Method: single particle / : Hjorth CK, McLellan JS

EMDB-43604: 
CCHFV GP38 bound to ADI-46152 and ADI-58048 Fabs
Method: single particle / : Hjorth CK, McLellan JS

PDB-8vww: 
CCHFV GP38 bound to ADI-46152 and ADI-58048 Fabs
Method: single particle / : Hjorth CK, McLellan JS

EMDB-29620: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG
EMDB-29621: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG
EMDB-29627: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG
EMDB-29628: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-29631: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-B)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG
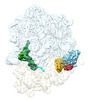
EMDB-29634: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

PDB-8fzd: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

PDB-8fze: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

PDB-8fzg: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

PDB-8fzh: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

PDB-8fzi: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-B)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

PDB-8fzj: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-45655: 
Cryo-EM structure of alpha5beta1 integrin in complex with NeoNectin
Method: single particle / : Werther R, Nguyen A, Estrada Alamo KA, Wang X, Campbell MG

EMDB-29618: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (State I-B)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-29619: 
ApoRF3 bound to an E. coli non-rotated ribosome termination complex, from focused classification and refinement (State I-B)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-29625: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (State II-A)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-29626: 
RF3-GDPCP bound to an E. coli non-rotated ribosome termination complex, from focused classification and refinement (State II-A)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-29629: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (State II-B)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-29630: 
RF3-GDPCP bound to an E. coli rotated ribosome, from focused classification and refinement (State II-B)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-29632: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (State II-C)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-29633: 
RF3-GDPCP bound to an E. coli rotated ribosome, from focused classification and refinement (State II-C)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG
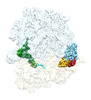
EMDB-29622: 
Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG
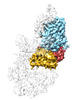
EMDB-29623: 
RF3-ppGpp bound to an E. coli rotated ribosome, from focused classification and refinement (State I-C)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG
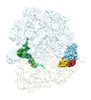
EMDB-29624: 
Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

PDB-8fzf: 
Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C)
Method: single particle / : Rybak MY, Li L, Lin J, Gagnon MG

EMDB-18438: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 1
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

EMDB-18439: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 2
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

EMDB-18440: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 3
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

EMDB-18443: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 4
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

EMDB-18460: 
mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 1
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

EMDB-18461: 
mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 2
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

PDB-8qrk: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 1
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

PDB-8qrl: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 2
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

PDB-8qrm: 
mt-SSU assembly intermediate in GTPBP8 knock-out cells, state 3
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

PDB-8qrn: 
mt-SSU in GTPBP8 knock-out cells, state 4
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

PDB-8qu1: 
mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 1
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

PDB-8qu5: 
mt-LSU assembly intermediate in GTPBP8 knock-out cells, state 2
Method: single particle / : Valentin Gese G, Cipullo M, Rorbach J, Hallberg BM

EMDB-18592: 
E.coli DNA gyrase in complex with 217 bp substrate DNA and LEI-800
Method: single particle / : Ghilarov D, Martin NI, van der Stelt M
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
